Examples
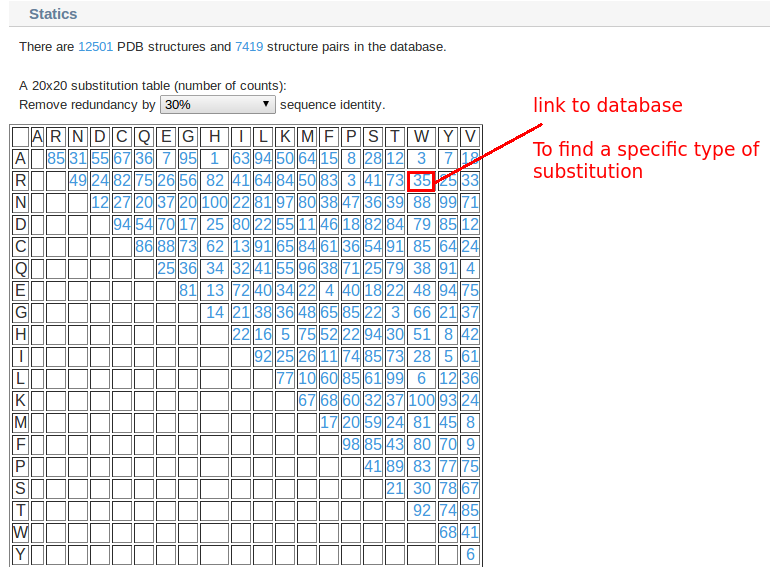
1. You can find all the data in the database in "data"
Data in the result table include:
1. Type: Substituted amino acid type
2. PDBs: Protein Data Bank id of the two proteins
3. Chains: PDB chain name
4. Lengths: Protein chain length
5. Positions: Substitution site according to residue sequence number in PDB file
6. H-bonds: Hydrogen Bond Number of protein backbone at substitution site according to STRIDE
7. ASAs: Solvent Accessible Surface Area at substitution site according to NACCESS
8. SSEs: Secondary structure of the two structures at substitution site according to DSSP
9. RMSD1 and RMSD2: RMSD of backbone atoms of the substituted residues (RMSD1) and the RMSD of the backbone atoms of flanking residues (RMSD2). RMSD1 measures the backbone perturbation of the substituted site, while RMSD2 evaluates the quality of the protein superimposition.
10. Plot: Ramachandran Plots of the substituted residues
11. View: Interactive 3D structure visualization of the two superimposed structures. (Please install java to view structures)
12. UniProt: Links to UniProt
13. Function: Information of function changes induced by single residue substitutions. It is obtained from the literatures of the respect crystal structures.
2. You can search the database for the PDB you are interested in "search"
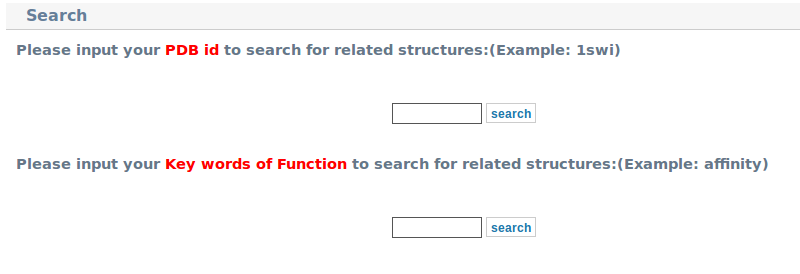
3. You can find all types of substitutions in "Statistics"